Millipede genomes reveal unique adaptations during myriapod evolution
Background
The Myriapoda, composed of millipedes and centipedes, is a fascinating but poorly understood branch of life, including species with a highly unusual body plan and a range of unique adaptations to their environment. Here, we sequenced and assembled two chromosomal-level genomes of the millipedes Helicorthomorpha holstii (assembly size = 182 Mb, N50 = 18.11 Mb mainly on 8 pseudomolecules), and Trigoniulus corallinus (assembly size = 449 Mb, N50 = 26.78 Mb mainly on 17 pseudomolecules).
Results
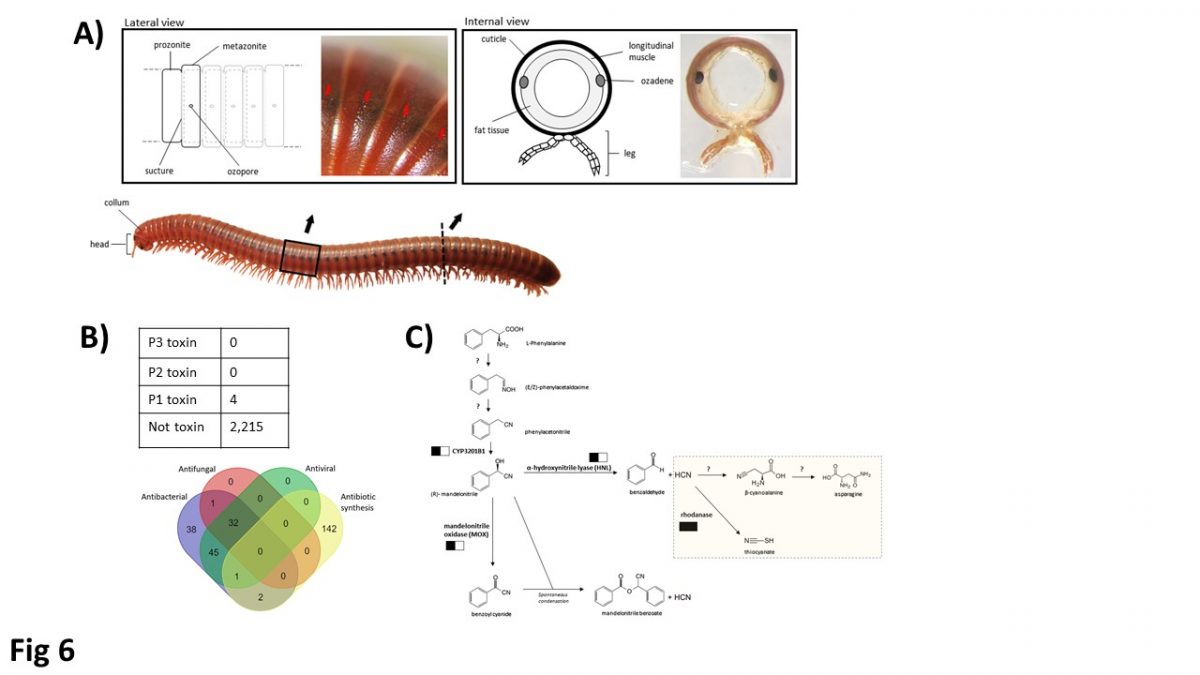
Unique genomic features, patterns of gene regulation, and defense systems in millipedes, not observed in other arthropods, are revealed. Both repeat content and intron size are major contributors to the observed differences in millipede genome size. Tight Hox, and the first loose ecdysozoan ParaHox homeobox clusters are identified, and a myriapod-specific genomic rearrangement including Hox3 is also observed. The Argonaute proteins for loading small RNAs are duplicated in both millipedes, but unlike in insects, an Argonaute duplicate has become a pseudogene. Evidence of post-transcriptional modification in small RNAs, including species-specific microRNA arm switching, providing differential gene regulation is also obtained. Millipedes possesses a unique ozadene defensive gland unlike the venomous forcipules found in centipedes. We identify sets of genes associated with the ozadene that play roles in chemical defense as well as anti-microbial activity. Macro-synteny analyses revealed highly conserved genomic blocks between the two millipedes and deuterostomes. Collectively, our analyses of millipede genomes reveal a series of unique adaptations have occurred in this major lineage of arthropod diversity.
Conclusions
The two high quality millipede genomes provided here shed new light on the conserved and lineage-specific features of millipedes and centipedes. These findings demonstrate the importance of the consideration of both centipede and millipede genomes, and in particular the reconstruction of the myriapod ancestral situation, for future research to improve understanding of arthropod evolution, and animal evolutionary genomics more widely.
Reference
- ^Qu Z, ^Nong WY, ^So WL, ^Barton-Owen T, ^Li YQ, Leung TC, Li C, Baril T, Wong AY, Swale T, Chan TF, Hayward A, Ngai SM, Hui JHL* . (accepted). Millipede genomes reveal unique adaptation of genes and microRNAs during myriapod evolution. PLOS Biology, 18(9), e3000636. (Link) (Story also covered in: Earth.com, EurekAlert, ScienceDaily, SCINEWS)
- Kenny NJ, Shen X, Chan TT, Wong NW, Chan TF, Chu KH, Lam HM, Hui JHL* (2015). Genome of the rusty millipede, Trigoniulus corallinus, illuminates diplopod, myriapod and arthropod evolution.Genome Biology and Evolution, 7(5), pp. 1280-1295.(Link) (Cover story of the journal)
- ECF – Millipede and Soil Biodiversity Citizen Science project, http://biodiversity.sls.cuhk.edu.hk/millipedes/