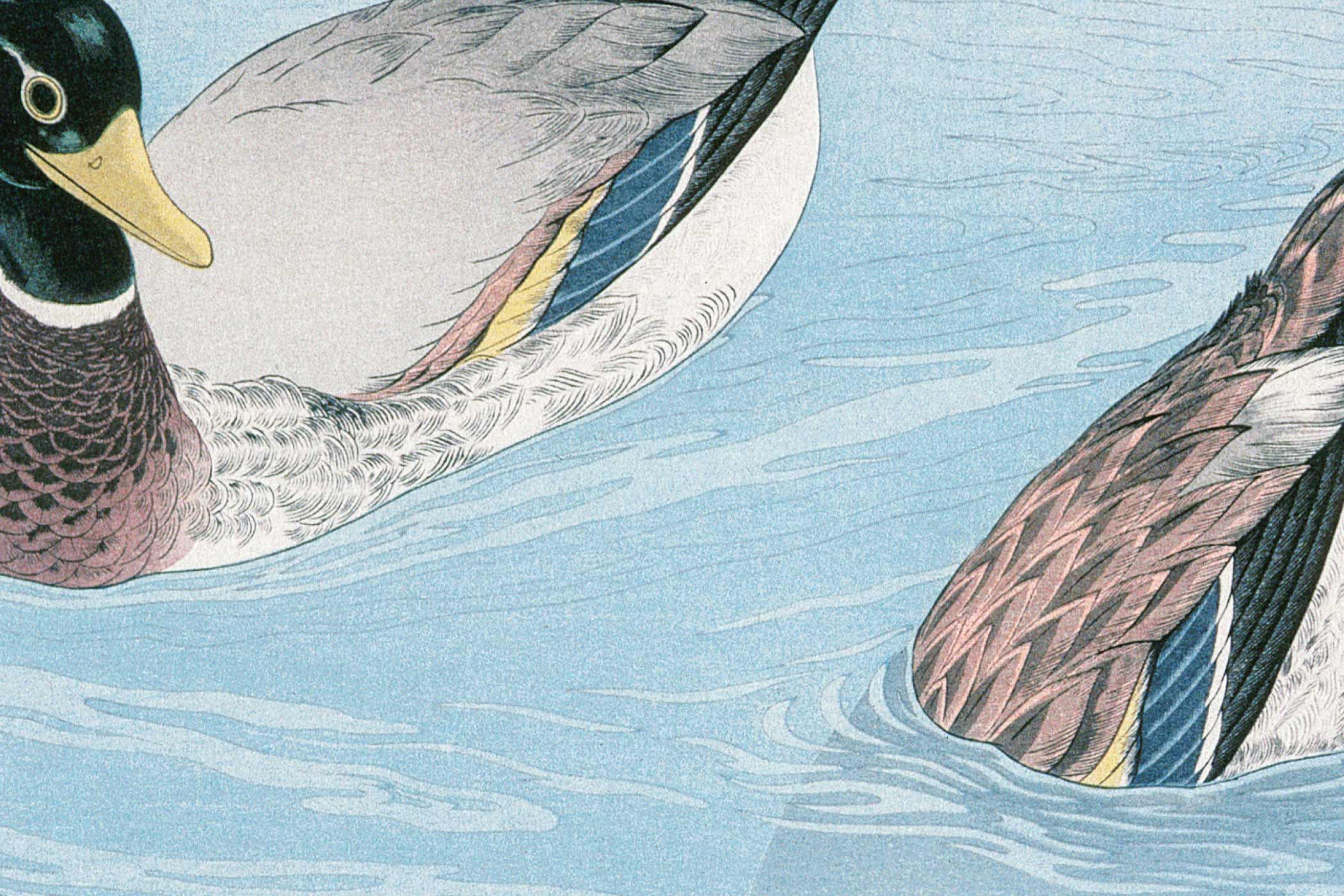
T. tridentatus 
C. rotundicauda
Whole genome duplication (WGD) has occurred in relatively few sexually reproducing invertebrates. Consequently, the WGD that occurred in the common ancestor of horseshoe crabs ~135 million years ago provides a rare opportunity to decipher the evolutionary consequences of a duplicated invertebrate genome. Here, we present a high-quality genome assembly for the mangrove horseshoe crab Carcinoscorpius rotundicauda (1.7Gb, N50 = 90.2Mb, with 89.8% sequences anchored to 16 pseudomolecules, 2n = 32), and a resequenced genome of the tri-spine horseshoe crab Tachypleus tridentatus (1.7Gb, N50 = 109.7Mb). Analyses of gene families, microRNAs, and synteny show that horseshoe crabs have undergone three rounds (3R) of WGD, and that these WGD events are shared with spiders. Comparison of the genomes of C. rotundicauda and T. tridentatus populations from several geographic locations further elucidates the diverse fates of both coding and noncoding genes. Together, the present study represents a cornerstone for a better understanding of the consequences of invertebrate WGD events on evolutionary fates of genes and microRNAs at individual and population levels, and highlights the genetic diversity with practical values for breeding programs and conservation of horseshoe crabs.
Data availability
The final genome assemblies have been deposited on NCBI with accession numbers WCHO00000000 and WCHN00000000, The raw reads generated in this study have been deposited to the NCBI database under the BioProject accession no. PRJNA574021 and PRJNA574023, for C. rotundicauda and T. tridentatus respectively. The genome annotation files are deposited in Figshare https://doi.org/10.6084/m9.figshare.13172414. All other data, if any, are available upon reasonable request.
Reference
- ^Nong WY, ^Qu Z, ^Li YQ, ^Barton-Owen T, ^Wong AY, Yip HY, Lee HT, Narayana S, Baril T, Swale T, Cao J, Chan TF, Kwan HS, Ngai SM, Panagiotou G, Qian PY, Qiu JW, Yip KY, Ismail N, Pati S, John A, Tobe SS, Bendena WG, Cheung SG, Hayward A, Hui JHL*. (2021). Horseshoe crab genomes reveal the evolutionary fates of genes and microRNAs after three rounds (3R) of whole genome duplication .Communications Biology, 4, 83. (Link)
- ^Qu Z, ^Leung TC, ^Nong WY, ^Yip HY, Lee IH, Cheung SG, Ngai SM, So WL, Bendena WG, Tobe SS, Hui JHL* . (2020). Hemolymph proteomics and gut microbiota of horseshoe crabs Tachypleus tridentatus and Carcinoscorpius rotundicauda. Frontiers in Marine Science (Link)
- Lee IH, Ng SY, Yip HY, Qu Z, Nong WY, Wong CF, Cheung SG*, Hui JHL* . (2020). An integrative approach to studying horseshoe crabs in Hong Kong: Field survey, Environmental DNA (eDNA) detection, and Citizen science platform. Book, Springer
- Field trail of juvenile horseshoe crab monitoring in Hong Kong using environmental DNA (eDNA) technique, http://biodiversity.sls.cuhk.edu.hk/horseshoecrab