Background
Homeobox-containing genes encode crucial transcription factors involved in animal, plant and fungal development, and changes to homeobox genes have been linked to the evolution of novel body plans and morphologies. In animals, some homeobox genes are clustered together in the genome, either as remnants from ancestral genomic arrangements, or due to coordinated gene regulation. Consequently, analyses of homeobox gene organization across animal phylogeny provide important insights into the evolution of genome organization and developmental gene control, and their interaction. However, homeobox gene organization remains to be fully elucidated in several key animal ancestors, including those of molluscs, lophotrochozoans and bilaterians.
Results
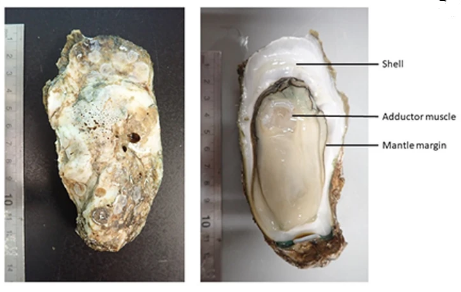
Here, we present a high-quality chromosome-level genome assembly of the Hong Kong oyster, Magallana hongkongensis (2n = 20), for which 93.2% of the genomic sequences are contained on 10 pseudomolecules (~ 758 Mb, scaffold N50 = 72.3 Mb). Our genome assembly was scaffolded using Hi-C reads, facilitating a larger scaffold size compared to the recently published M. hongkongensis genome of Peng et al. (Mol Ecol Resources, 2020), which was scaffolded using the Crassostrea gigas assembly. A total of 46,963 predicted gene models (45,308 protein coding genes) were incorporated in our genome, and genome completeness estimated by BUSCO was 94.6%. Homeobox gene linkages were analysed in detail relative to available data for other mollusc lineages.
Conclusions
The analyses performed in this study and the accompanying genome sequence provide important genetic resources for this economically and culturally valuable oyster species, and offer a platform to improve understanding of animal biology and evolution more generally. Transposable element content is comparable to that found in other mollusc species, contrary to the conclusion of another recent analysis. Also, our chromosome-level assembly allows the inference of ancient gene linkages (synteny) for the homeobox-containing genes, even though a number of the homeobox gene clusters, like the Hox/ParaHox clusters, are undergoing dispersal in molluscs such as this oyster.
Availability of data and materials
The final chromosome assembly was submitted to NCBI Assembly under accession number WFKH00000000 in NCBI. The raw reads generated in this study have been deposited to the NCBI database under the BioProject accessions: PRJNA576886, the genome annotation files were deposited in the Figshare https://doi.org/10.6084/m9.figshare.12715490.
References
1. ^Li YQ, ^Nong WY, ^Baril T, Yip HY, Swale T, Hayward A*, Ferrier DE*, Hui JHL* . (2020). Reconstruction of ancient homeobox gene linkages inferred from a new high-quality assembly of the Hong Kong oyster (Magallana hongkongensis) genome. BMC Genomics, 21, 713.(Link)